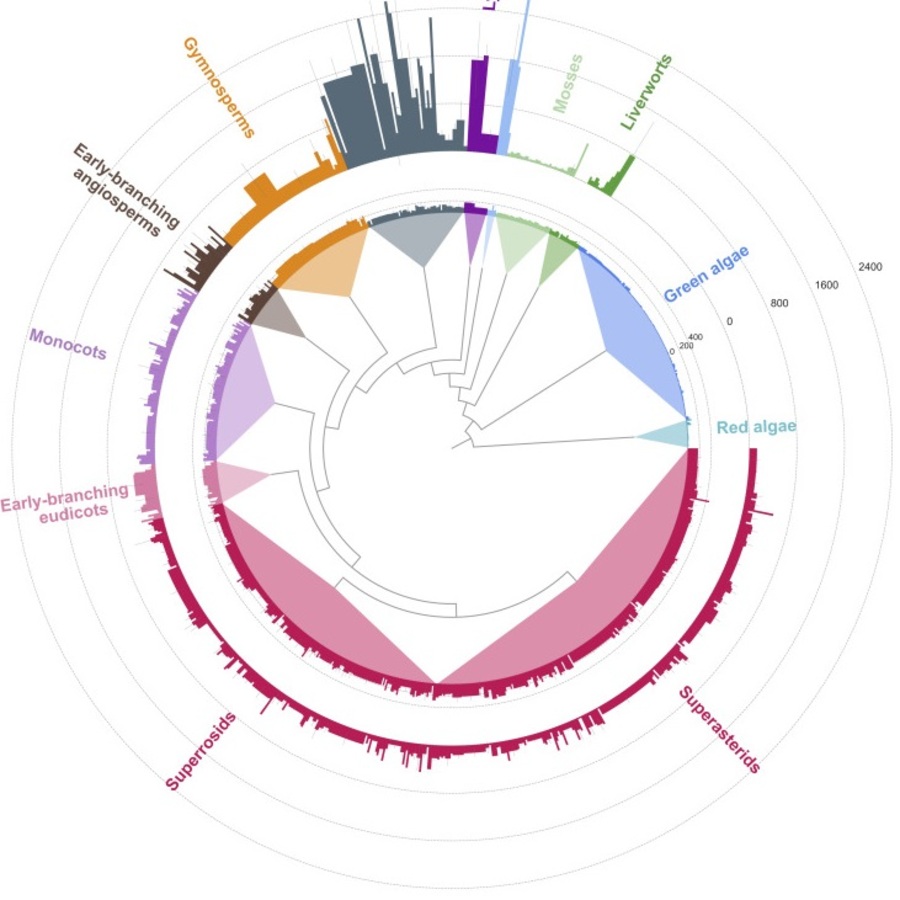
PEB scientists working alongside scientists from the University of Bonn in Germany have analysed a massive dataset of transcriptomes from over a thousand land plant species to expand the understanding of the pentatricopeptide repeat (PPR) protein superfamily in RNA editing.
PPR proteins are eukaryotic RNA binding proteins with diverse functions in RNA processing, and act as regulators of post-transcriptional control in organelles. A subfamily of PPR proteins are involved in RNA editing, a process where targeted cytidine bases are converted to uridines. This process results in changes in RNA sequences compared to the DNA sequence, which can produce a different functional product.
The study discovered a high variability in the size of the PPR superfamily throughout the plant kingdom, alongside clade specific expansion of the RNA editing subfamily and variations within their RNA binding repeat modules. These variations correlated with the expansion of another family of editing factors that act as molecular chaperones to enhance targeted RNA binding.
The results also showed key differences in the domain believed to catalyse the editing process itself, hypothesised to be responsible for the process of uridine to cytidine RNA editing. The large number and diversity of sequences allowed for the development of a high confidence tertiary model of the editing domain structure, which supports the hypothesis of its enzymatic function.
Deeper understanding of the important PPR superfamily has a crucial impact on the development of biotechnological tools for targeted RNA editing.
Gutmann, B., Royan, S., Schallenberg-Rüdinger, M., Lenz, H., Castleden, I. R., Mcdowell, R., Vacher, M. A., Tonti-Filippini, J., Bond, C. S., Knoop, V. and Small, I. D. (2020) The Expansion and Diversification of Pentatricopeptide Repeat RNA-Editing Factors in Plants Molecular Plant, 13(2), pp. 215-230.
rQZfM5kcbmYMReygY