Research databases to aid new discoveries
PEB has built a number of valuable research databases across Australia that are available to researchers in several formats, from GUI interfaces for queries, to data downloads and web services.
Computational Biology Tools
PEB uses in-house or has specialised a range of tools for proteomic and metabolomic identification, and nucleotide or protein sequence analysis and alignment.
Research Applications and Databases
 APP
https://webapps.plantenergy.uwa.edu.au/APP/
The Arabidopsis Proteotypic Predictor (APP) takes a Arabidopsis
Gene Initiative (AGI) identifier (eg At3g15020.1) or the amino
acid sequence of an Arabidopsis protein, performs an in silico
trypsin digestion and provides details of all resulting
peptides.
APP
https://webapps.plantenergy.uwa.edu.au/APP/
The Arabidopsis Proteotypic Predictor (APP) takes a Arabidopsis
Gene Initiative (AGI) identifier (eg At3g15020.1) or the amino
acid sequence of an Arabidopsis protein, performs an in silico
trypsin digestion and provides details of all resulting
peptides.
 MetAlign
https://webapps.plantenergy.uwa.edu.au/MetAlign
Transform AMDIS data files. Useful in metabolomics
research.
MetAlign
https://webapps.plantenergy.uwa.edu.au/MetAlign
Transform AMDIS data files. Useful in metabolomics
research.
 PPR
https://ppr.plantenergy.uwa.edu.au
446262 Pentatricopeptide repeat proteins from 1121 species. The
RNA-binding pentatricopeptide repeat (PPR) family comprises
hundreds to thousands of genes in most plants, but only a few
dozen in algae, evidence of massive gene expansions during land
plant evolution.
PPR
https://ppr.plantenergy.uwa.edu.au
446262 Pentatricopeptide repeat proteins from 1121 species. The
RNA-binding pentatricopeptide repeat (PPR) family comprises
hundreds to thousands of genes in most plants, but only a few
dozen in algae, evidence of massive gene expansions during land
plant evolution.
 Chloe
https://chloe.plantenergy.edu.au
Chloe is a website when you can annotate your organelle DNA
sequences from fasta/genbank files. It also offers the ability
to search and compare other organelles and even reannotate from
an NCBI Id.
Chloe
https://chloe.plantenergy.edu.au
Chloe is a website when you can annotate your organelle DNA
sequences from fasta/genbank files. It also offers the ability
to search and compare other organelles and even reannotate from
an NCBI Id.
 Thermo Scientific Proteome Discoverer Node
wheatproteome.org/proteover
Thermo Scientific Proteome Discoverer Node (plugin) to analyse
protein turnover using isotopically labelled data.
Thermo Scientific Proteome Discoverer Node
wheatproteome.org/proteover
Thermo Scientific Proteome Discoverer Node (plugin) to analyse
protein turnover using isotopically labelled data.
 Wheatproteome
https://wheatproteome.org
This resource provides a range of ‘omics data types in an
easily accessible format currently based around the 2014 IWGSC
Triticum aestivum chromosome assembled draft genome
release.
Wheatproteome
https://wheatproteome.org
This resource provides a range of ‘omics data types in an
easily accessible format currently based around the 2014 IWGSC
Triticum aestivum chromosome assembled draft genome
release.
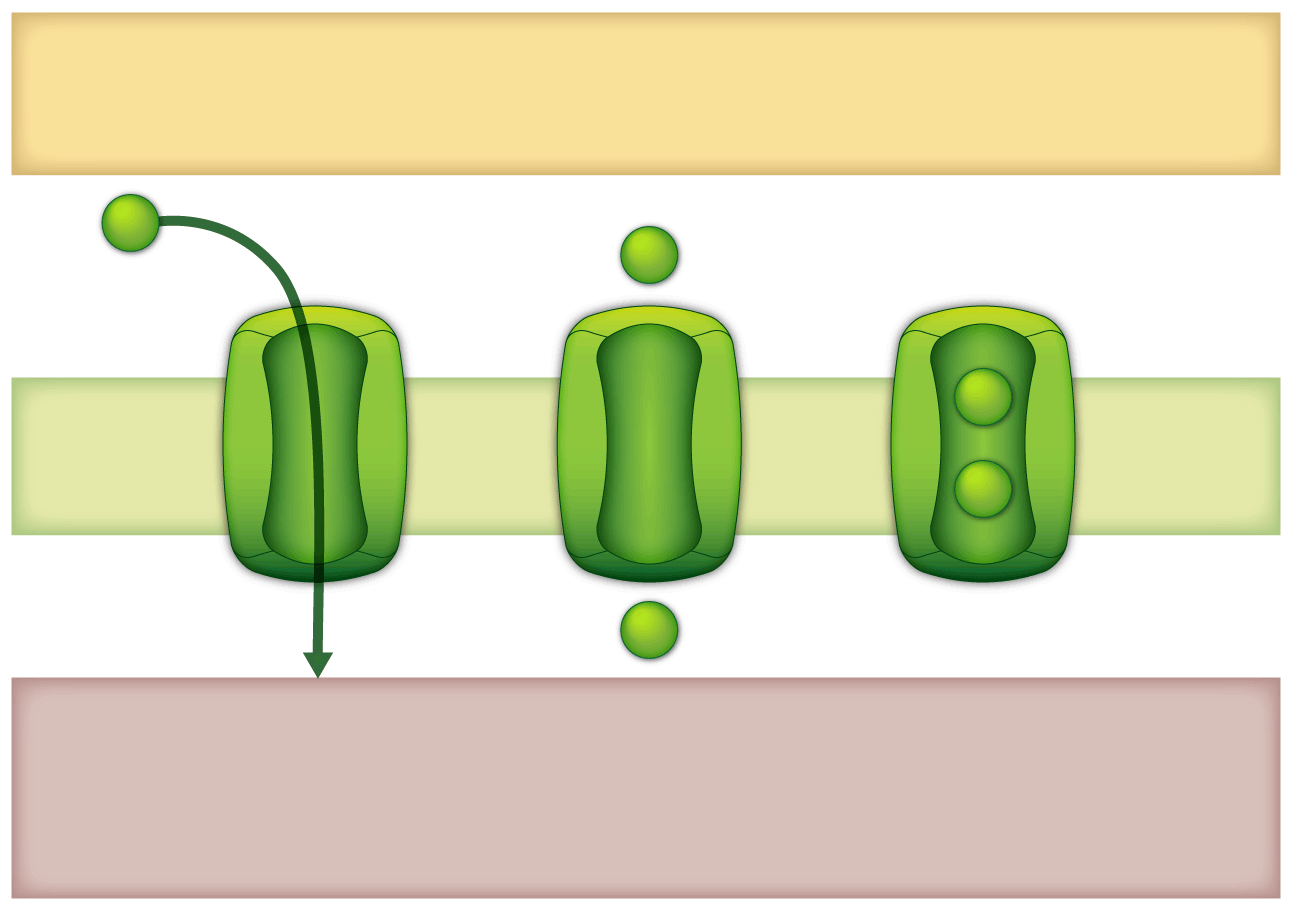 Mitochondrial Protien Import Components (MPIC)
https://webapps.plantenergy.uwa.edu.au/applications/mpic/
MPIC provides a simple and centralised resource for all
components of the mitochondrial protein import apparatus to
provide insight into each gene/protein function(s) throughout
evolution.
Mitochondrial Protien Import Components (MPIC)
https://webapps.plantenergy.uwa.edu.au/applications/mpic/
MPIC provides a simple and centralised resource for all
components of the mitochondrial protein import apparatus to
provide insight into each gene/protein function(s) throughout
evolution.
 SubCellular Proteomic Database (SUBA)
https://suba.live
The SubCellular Proteomic Database (SUBA) houses large scale
proteomic and GFP localisation sets from cellular compartments
of Arabidopsis.
SubCellular Proteomic Database (SUBA)
https://suba.live
The SubCellular Proteomic Database (SUBA) houses large scale
proteomic and GFP localisation sets from cellular compartments
of Arabidopsis.




